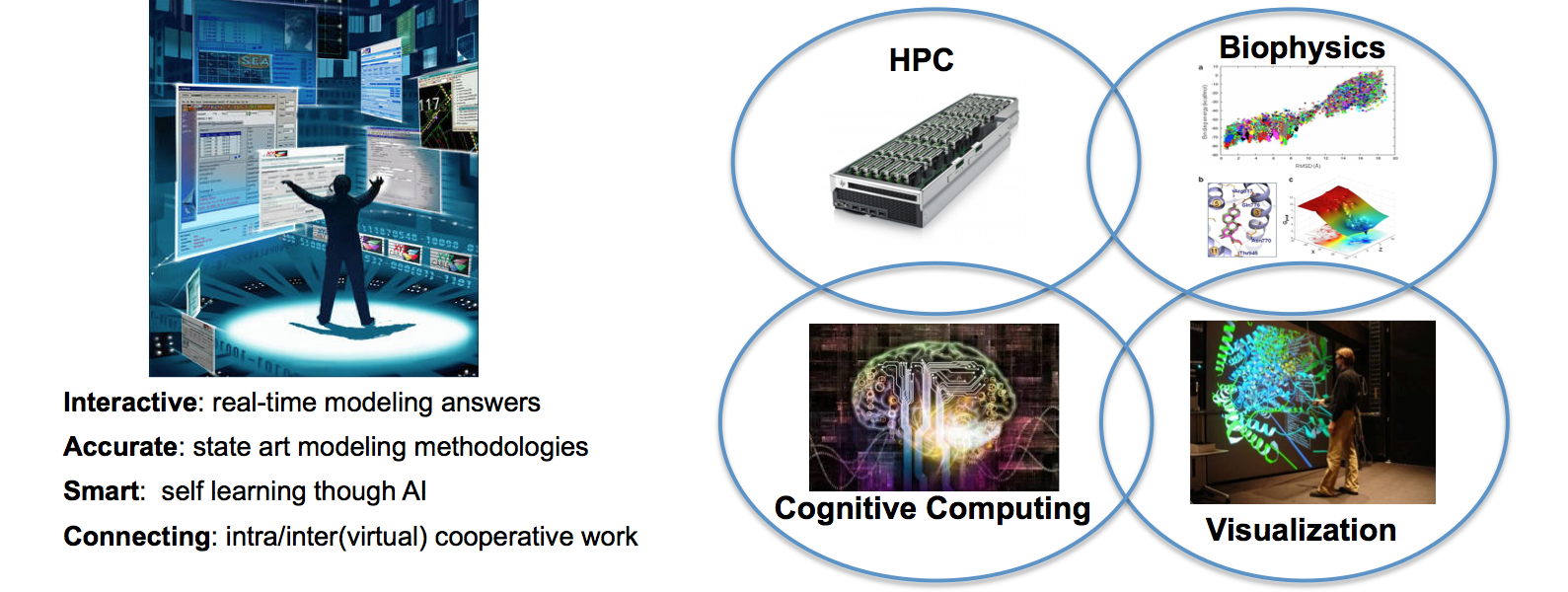
Possibly our most ambitious project, designing the next generation smart and interactive drug design software, by combining biophysical modeling, advanced graphics (and virtual reality), high performance computing and cognitive computing.
Summary
Computer simulations are increasingly taking over in design processes. Still, complex design procedures, such as biomolecular engineering (drug design, protein engineering, etc.), largely benefit from human expertise. We aim at building a future where smart and quick algorithms will work together, in real time, with human intuition. Moreover, the software will learn from such interaction, and improve in automatic predictions further nurturing human creativity. Imagine a drug design team performing interactive simulations, introducing drug changes and checking the effects -via simulation- in few seconds. They work in a virtual reality environment, connected worldwide, stimulating their insight and creating drugs. Computer simulation and visualization spark human intuition that, in turn, feeds new simulations teaching the software to improve its automatic procedures. We are working on such breakthrough technological platform by introducing advances in four interdisciplinary research areas:
- In biomolecular modeling where our state-of-the-art techniques (PELE) will allow for a fast and robust prediction of molecular interactions.
- In HPC hardware, where number of cores and presence of accelerators, such as GPU or FPGA, will turn the next generation workstations to a nowadays small supercomputer.
- In visualization, providing richer perception and introducing virtual (or augmented) realities.
- In Cognitive Computing, extracting knowledge from multiple and heterogeneous sources, together with algorithm evolution based on usage and success/error analysis.
Objectives
Current ongoing work involves:
- Designing the overall interface, from a User Experience (UX) point of view (see, for example https://youtu.be/1OIYkChsOl8)
- Connecting the biophysical simulation code with HPC, allowing real-time steering of the simulations (see for example the MontBlanc project).
- Developing enhanced molecular visualization and virtual reality tools (see for example IEEE transactions on visualization and computer graphics (2016) 22:718-727)
- Developing a machine learning based assistant that learns from simulations and user actions, and suggests more effective simulations to the user. Work in this line centers on Adaptive Sampling Techniques and Consensu Scorung Functions.
