Molecular Modeling
-

Advanced docking strategies
Molecular recognition is the basis of biological function. Docking is among a number of structural bioinformatics strategies trying to understand molecular recognition, both to understand structure to function relationships in macromolecules, and to derive tools useful in fields like drug design.
-

Docking for interactomics and personalized medicine
The ultimate goal of our protein-protein docking tools and their application to the characterization of protein interactions of therapeutic interest is to contribute towards personalized medicine and drug discovery targeting protein-protein interactions.
-
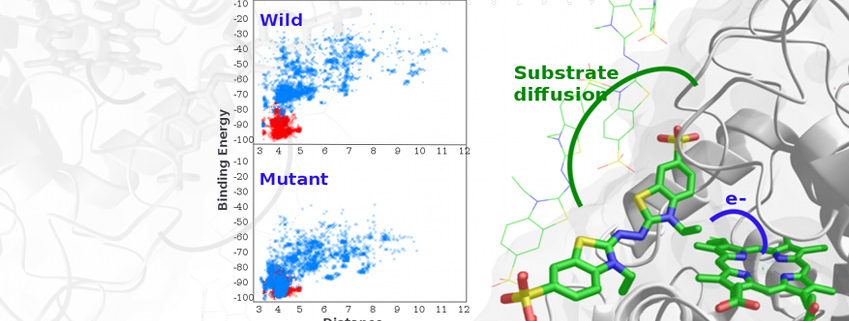
Enzymatic Catalysis and Protein Engineering
As one of our main research lines, we model enzymatic activity addressing not only the atomic (and electronic) detailed mechanism but also suggesting mutations for changes in activity, substrate specificity, etc.
-

New approaches to the prediction of protein structure and interactions
The combined exploration of the sequence and structure spaces with large scale simulations leads to a new generation highly accurate prediction methods useful in applications such as the design of new drugs.
-

PELE: Monte Carlo methods for protein/DNA-ligand interactions
A key line of research in the lab is the development of PELE (Protein Energy Landscape Exploration), a fast and accurate Monte Carlo software for mapping protein(and DNA)-ligand interactions.
-

Software development for protein-protein interactions
The main focus of our research is the development and optimization for High-Performing-Computing (HPC) of computational methodology for the structural and energetic characterization of protein interactions at molecular level.
