The new computational method, published in Nature Communications, could help design new proteins with different biotechnological and biomedical applications
AlphaFold2, the artificial intelligence (AI) tool, developed by DeepMind from Google, that can predict accurate models of proteins, the fundamental components of cellular systems, has led to a revolution that has benefited different fields of science, from drug design to the study of evolution.
Now, a team of researchers at the Barcelona Supercomputing Center - Centro Nacional de Supercomputación (BSC-CNS) has taken advantage of AlphaFold2's ability to predict the structures of the thousands of proteins that constitute different protein families to develop a computational method that analyses the degree of conservation throughout evolution - and therefore their importance - of regions critical to the structure and function of each protein family (known as energetically frustrated regions).
The new computational method and the results obtained contribute to the basic understanding of the evolution of proteins and could help in the design of new ones with different biotechnological and biomedical applications.
The scientists, who are part of the BSC's Life Sciences department directed by ICREA Prof. Alfonso Valencia, have also benefited from the great computational capacity of the MareNostrum supercomputer to carry out this study, which has been led by Dr. Gonzalo Parra, from the Computational Biology group. The results of this research, in collaboration with groups from Argentina, Chile and the United States, have been published in the journal Nature Communications.
"These new conditions allow us, for the first time, to tackle big questions about the relationship between the three-dimensional structure of proteins and their functional capacities, with evolutionary conservation and protein structures as a guide. A subject that combines scientific interest with direct implications in biotechnology for the rational design of proteins with new properties," says Alfonso Valencia.
Epidemiological surveillance of emerging pathogens
The recent coronavirus pandemic highlighted the importance of understanding the evolutionary mechanisms that operate over pathogens with potential infectivity in humans. For this reason, one of the cases that has been studied in detail in this study is that of SARSCov2 proteins, which are still poorly characterised in terms of their functional properties. All the proteins encoded by the SARSCov2 genome were analysed in the context of the proteins encoded by the hundreds of known coronavirus genomes, both those that infect humans as well as other species.
Analyses on the PLPro protein, a protein of direct interest for antiviral development, showed that the evolutionary history of the 124 related coronaviruses can provide many clues as to how they adapt to their hosts and improve their infectivity strategies. "In particular, catalytic site changes were detected that are potentially responsible for virus adaptation to different hosts, opening new doors for the development of compounds targeting these sites," says Valencia.
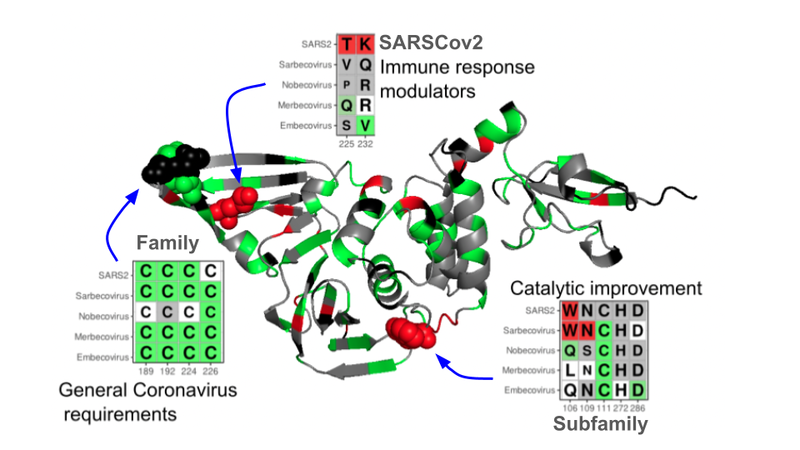
Figure: Energetic frustration map of PLPro where evolutionary constraints at different levels (all coronaviruses, sarbecoronaviruses, SARSCov2) are shown. In green regions that are important for stability, in red regions that are important for function. The most important regions are highlighted.
This analysis required less than a week of computation on the BSC's MareNostrum supercomputer. Similar strategies can be applied in the future to analyse the evolution of new pathogens of epidemiological interest as soon as their genomes become available. Such analyses could facilitate the development of vaccines or other strategies to fight such viruses.
Reference: "Local energetic frustration conservation in protein families and superfamilies" https://www.nature.com/articles/s41467-023-43801-2
Picture: BSC Computational Biology group

