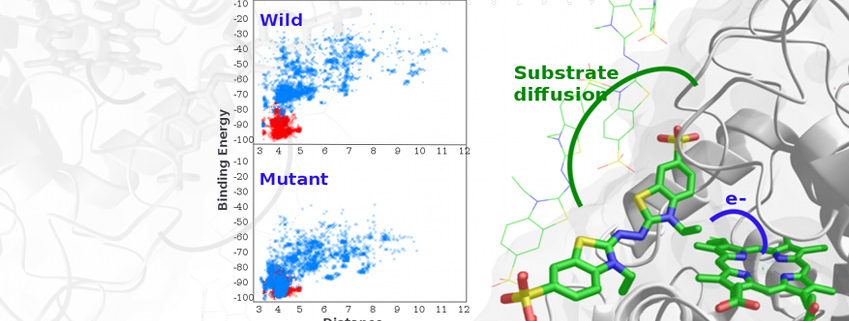
As one of our main research lines, we model enzymatic activity addressing not only the atomic (and electronic) detailed mechanism but also suggesting mutations for changes in activity, substrate specificity, etc.
Summary
Combining computational biophysics and biochemistry, we study enzymatic mechanisms at a detailed atomic and electronic level. Biophysical techniques, mainly molecular dynamics (MD) and our in-house Monte Carlo sampling technique (PELE), model ligand migration (and binding), addressing substrate-enzyme recognition and induced fit. Biochemical predictions, centered on applying quantum chemistry techniques (mainly through mixed QM/MM methods), aim to map the chemical process.
Studies have involved characterisation of multiple enzymatic mechanisms (oxidases, methyltransferases, esterases, etc.). Moreover, we have focused on understanding the effects of several mutations in modulating enzymatic activity.
More recently we have turned our attention to in silico rational and directed evolution, where we aim to predict new mutations towards adding specificity or enhanced activity for a substrate. In a recent study, for example, by engineering two surface histidine residues, we turned an ABTS inactive manganese peroxidase into an active one (ACS Catalysis (2016) 6:1624-1629).
Most of these studies are conducted through collaborations with experimental labs and industry, including Angel Martinez (CSIC-CIB), Miguel Alcalde (CISC-ICP), Tom Spiro (University of Washington), Joseph Jez (Washington University), NOVOZYMES, etc.
Objectives
Objectives mainly involve:
- Mapping, at an atomic and electronic level, enzymatic mechanisms.
- Rationalisation of mutational effects into the enzymatic mechanism
- Prediction of new mutations towards adding specificity and activity

