 Speaker: Ross C. Walker, Associate Professor,
Speaker: Ross C. Walker, Associate Professor,
San Diego Supercomputer Center & Department of Chemistry and Biochemistry,
University of California San Diego
Abstract: This talk will focus on the impact that GPUs have had on Molecular Dynamics (MD) Simulations. In particular it will highlight the massive performance improvements that GPUs have brought to MD simulations with AMBER. Individual desktops with NVIDIA Kepler based GPUs can routinely provide simulation rates (for a 25,000 atom simulation) exceeding 130ns/day per GPU. A single desktop is now able to provide aggregate performance exceeding half a microsecond a day. Employing various tricks for longer time steps and constrained degrees of freedom and performance can exceed microseconds per day. Meanwhile replica exchange approaches to accelerating convergence enable hundreds of GPUs to be employed in parallel
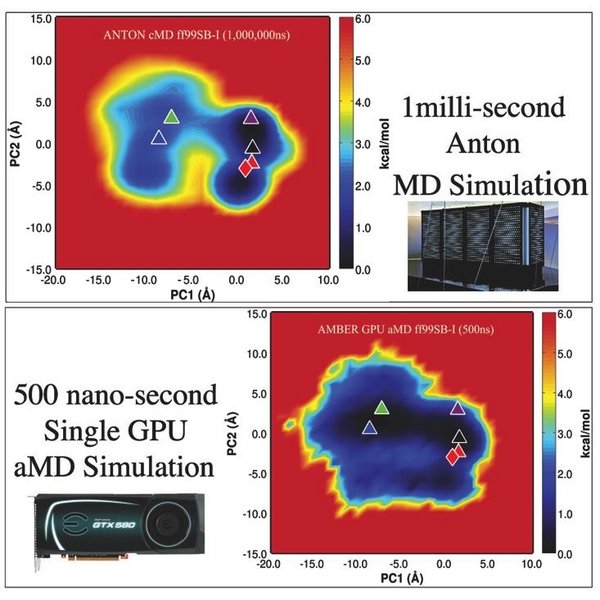
The GPU revolution has transformed the MD landscape. No longer is access to supercomputer resources required to routinely access microsecond timescales and beyond. The world of MD research is now flat, with all researchers, young and old, rich and poor being able to run simulations that previously were restricted to those privileged enough to have routine access to supercomputers. Further, recent focus on algorithmic improvements aimed at accelerating the rate at which phase space is sampled has transformed things further. A recent success, which will be highlighted here, has been the reproduction and extension of key results from the DE Shaw 1 millisecond Anton MD simulation of BPTI (Science, Vol. 330 no. 6002 pp. 341-346) with just 5 days of dihedral boosted AMD sampling on a single GPU workstation, (Pierce L, Walker R.C. et al. JCTC, 2012, p5092). These results show that with careful algorithm design it is possible to obtain sampling of rare biologically relevant events that occur on the millisecond timescale using just a single $500 GTX680 Graphics Card and a desktop workstation.
