Cancer genomics
Our main interest and activity around Cancer Genomics is to identify somatic genomic variations (mutations) that are responsible for the development and progression of tumours. For this, we generate and apply different bioinformatic methodologies.
Summary
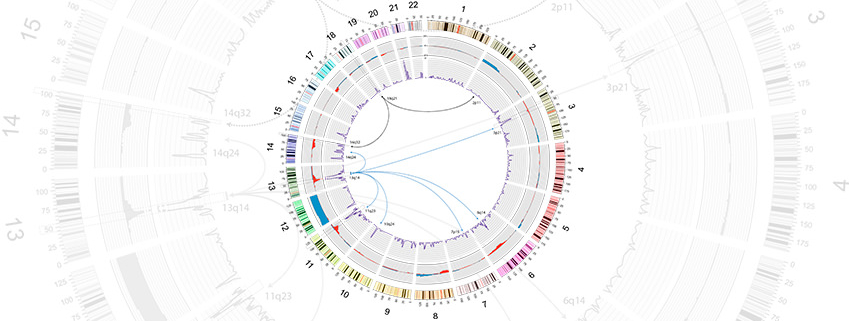
Cancer genomics covers a large part of our interests and research activity. Within various international consortia, we are developing and applying analysis tools and approaches to shed light on the underlying genomic and functional basis of several types of tumours. In the race to understand the biology of tumours and to find particular and personalised therapies, our contribution to the field is centred on the computational analysis of variation at different levels. In this context, we have recently developed SMuFin (for Somatic MUtation FINder), a reference-free method for the identification of genomic mutations that are responsible for the development and progression of tumours.
By directly comparing the sequences of normal and tumour cells, SMuFin can identify (in a single run) nearly all types of somatic variants, from single nucleotide variations, to large structural changes implying complex rearrangements of chromosomes, e.g. those involved in chromotripsis and chomoplexy. With this and other tools in hand, we aim to search for complex genomic landscapes that are associated with cancer and to identify their underlying mechanisms. We are also interested in uncovering, beyond the affected genes, to what extent the variation in gene regulatory regions can lead to oncogenesis. In this context, we will also use several tools developed in the group for the identification of promoters and enhancers in eukaryotic genomes (e.g. ReLA ). The combination of the identification of variants associated with tumour formation and the analysis of regulatory regions can allow us to find this missing link between gene regulation and cancer origin and progression.
Objectives
- Application of the tools for answering specific biomedical questions, and given the potential of these tools, we also continue improving and developing further these methodologies in different directions.
